Analysis of Cell Viability (Fe)
¡Los gráficos interactivos pueden tardar en cargar!
Key Findings and Interpretation
The regression model for cell viability (Fe) is statistically significant (P-Value = 0.000) and explains a large portion of the data's variability, with an R-sq of 82.31%. The model is complex, involving numerous significant two-way and three-way interactions, indicating that the components' effects are highly interdependent.
The most influential terms are the interactions Nano*Spiruline (P=0.000) and Water*Spiruline (P=0.000), both of which have a strong negative impact on cell viability. The interactions Alginate*Spiruline (P=0.005) and Spiruline*Calcium (P=0.007) are also highly significant. It is critical to note that the model shows a significant Lack-of-Fit (P=0.000), suggesting that the model may not fully capture the complex underlying relationships. Furthermore, the large difference between R-sq (82.31%) and R-sq(pred) (53.86%) indicates the model may be over-fit and might not be as effective at predicting new, unseen data.
Regression Equation
The relationship between the components and the cell viability response is described by the following equation, where the process variable Calcium is coded (-1 for 1%, +1 for 5%):
Model Goodness-of-Fit
The statistical model provides a strong fit to the experimental data, but with some caveats. The key metrics from the "Model Summary" table are:
- R-sq = 82.31%: The model explains over 82% of the variation in the cell viability data, indicating a strong correlation.
- R-sq(adj) = 76.62%: The adjusted R-squared remains high, suggesting the model is not unnecessarily complex.
- R-sq(pred) = 53.86%: The predicted R-squared is considerably lower than the R-squared value. This suggests caution should be used when using the model to predict new outcomes, as its predictive power is moderate.
- S = 0.0887339: The standard error of the regression is relatively low.
While the overall regression is highly significant (P-Value = 0.000), the significant Lack-of-Fit is a critical finding that warrants further investigation or model refinement.
Model Summary: Stepwise Selection
The following table shows the stepwise selection process for the final model. The last row, highlighted, represents the chosen model with the best combination of explanatory and predictive power.
| Step | S | R-sq (%) | R-sq(adj) (%) | R-sq(pred) (%) |
|---|---|---|---|---|
| 1 | 0.148132 | 40.14 | 34.85 | 16.24 |
| 2 | 0.123709 | 59.48 | 54.57 | 38.74 |
| 3 | 0.109708 | 69.10 | 64.27 | 49.28 |
| 4 | 0.100592 | 74.83 | 69.96 | 53.73 |
| 5 | 0.0976898 | 77.03 | 71.67 | 52.68 |
| 6 | 0.0948191 | 79.08 | 73.31 | 52.39 |
| 7 | 0.0887339 | 82.31 | 76.62 | 53.86 |
Model Diagnostic Plots
To ensure the validity of the statistical model, a series of diagnostic plots were generated. These plots help confirm that the assumptions of the regression analysis are met. Below is a guide to interpreting each plot:
- Normal Probability Plot: This plot checks if the residuals are normally distributed. For a perfect normal distribution, the data points would align perfectly along a straight diagonal line. The goal is to see our experimental points fall closely to this theoretical line. Significant deviations may indicate that the assumption of normality is not met.
- Residuals vs Fits: This plot is used to detect non-constant variance, missing terms, or outliers. The points should be randomly scattered around the horizontal line at zero. Any clear pattern, such as a curve or a funnel shape, would suggest a problem with the model.
- Histogram of Residuals: This provides another visual check for the normality of residuals. The distribution should be roughly symmetric and bell-shaped, centered around zero.
- Residuals vs Order: This plot helps to verify that the residuals are independent of one another. The data points should show no discernible trend or pattern. Any systematic pattern could suggest that the order of the experiments influenced the results.
Pareto Chart of Effects
The Pareto chart visually ranks the importance of each factor and interaction on the cell viability response. The red line indicates the threshold for statistical significance (α=0.05). Effects that cross this line are considered the most influential drivers of the process.
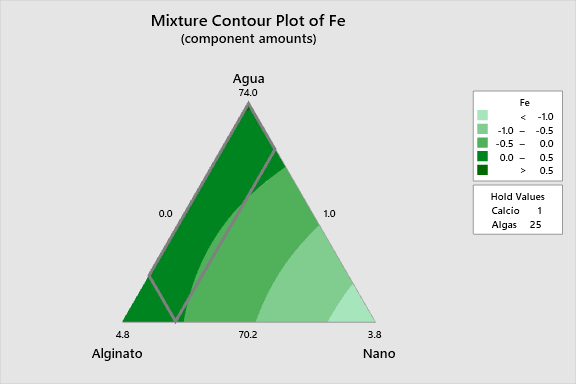
2D Contour Plots
The following interactive 2D contour plots show how pairs of variables influence cell viability while holding the other factors at constant levels. These maps are essential for identifying optimal regions in the formulation space.
3D Surface Plots
These interactive 3D plots provide an intuitive view of the response surface. Each colored surface represents the predicted cell viability response based on the model for a specific combination of held factors.
Overlaid on the surfaces are the data points from the actual experiments. The solid dots (●) represent the actual, measured viability values, while the crosses (+) show the values predicted by the model for those same experimental conditions. The vertical distance between a dot and its corresponding cross represents the residual error for that point. A good model will have these points lying close to the surface, indicating small errors.